RESEARCH
Group description and research lines.
Epigenetics is the term used in biology to refer to chromatin structure and DNA modifications that are stable over rounds of cell division but do not involve changes in the DNA sequence. Epigenetics play a pivotal role in cellular differentiation, allowing cells to stably maintain different characteristics despite containing the same genomic DNA. Epigenetic processes are also involved in gene silencing, X chromosome inactivation, reprogramming and are thought to be one of the major limitations to cloning. One of the main interests of this group is Genomic Imprinting. Imprinted genes are expressed from only one parental allele, the other is silenced by epigenetic modifications, classically involving DNA methylation and asymmetric chromatin structure. Imprinted genes are typically involved in embryonic growth and development. Abnormal imprinted gene expression is one of the most frequent aberrations in carcinogenesis.
Imprinting group

X
![]()
The aims of our group include learning about the control mechanisms involved in genomic imprinting, and how genetic and epigenetic variation in imprinted genes is associated with cancer susceptibility and fetal development. Some of our more specific research goals include:
- Identifying novel imprinted loci in humans
- Genome-wide methylation profiling during human development to understand the processes involved in imprint protection
- Profiling methylation aberrations in patient with classical imprinting disorders
- Epigenetic characterization of the placenta and its relationship to intrauterine growth restriction and other pregnancy associated-disorders
- Understand the role of imprinted genes in both childhood and adult tumours
SELECTED ARTICLES
(last update: 31th July 2023)
SUPPLEMENTAL ITEMS
Informations supplementing publications.
Imprinting in the human placenta
Compared with somatic tissues, the cells of the placenta have a unique epigenetic profile that dictates its transcription patterns, which when disturbed may be associated with adverse pregnancy outcomes. One major class of genes that is dependent on strict epigenetic regulation in the placenta are those subject to genomic imprinting. Following on from the discovery of the first placenta-specific DMR in humans, the C19MC, many recent studies have identified many genes subject to placenta-specific imprinting. The data available here complements our publications and supplementary information giving details of the precise location of human placenta-specific DMRs.

Names and locations of placenta-specific maternal transient DMRs
Placenta 5hmC profile
The recent discovery that 5mC can be oxidised to 5 hydromethylcytosine (5hmC) by TET proteins has revealed the "sixth base" of DNA and provides additional complexity to what was original thought to be a stable repressive mark. However the genome-wide distribution of 5hmC in different tissue is currently limited. Here we sought to define loci enriched for 5hmC in the placenta genome by combining oxidative bisulphite (oxBS) treatment with high-density Illumina Infinium HM450k methylation arrays and confirming our observations using hMeDIP-seq and T4 β-glucosyltransferase (T4-BGT) assays. Furthermore we compare our results from placenta with published datasets in brain (Lunnon et al., Genome Biol 2016). Despite identifying over 17,000 high-confidence loci with consistent 5hmC enrichment in placenta, this mark is relatively sparse when compared to cerebellum and frontal cortex. Here you can survey the top 500 "BumpHunter" defined intervals for placenta, cerebellum and frontal cortex. Surprisingly we identify 5hmC at numerous imprinted loci, often overlapping regions associated with parent-of-origin allelic 5mC in a tissue-specific fashion. This suggests that 5hmC should be considered when characterizing some imprinted DMRs using techniques that do not distinguish 5mC from 5hmC in placenta and brain.

Top 500 "bumps" of 5hmC for placenta

Top 500 "bumps" of 5hmC for cerebellum

Top 500 "bumps" of 5hmC for cortex
Cancer selection
It has been postulated imprinting aberrations are a common occurrence in cancer with some imprinted methylation signatures proposed as a risk marker in solid tumours. To gain a better understanding of the role of imprinting in cancer we have characterized both copy number and methylation in over 280 cancer cell lines from the Catalogue Of Somatic Mutations In Cancer (COSMIC) repository and confirm our observations in primary tumour datasets from The Cancer Genome Atlas (TCGA). The data available here represent the supplementary information from our publications characterising imprinting signatures in cancer.

Circos graphs for average methylation and copy-number at imprinted DMRs in cancer cell lines

Extent of copy-number aberrations encompassing imprinted loci in cancer cell lines and TCGA primary cancer samples
Nomeculature of imprinted DMRs and their precise locations
To date, primary methylation defects of some well characterised imprinted DMRs are directly responsible for imprinting disorders including Beckwith-Wiedemann syndrome (BWS; OMIM 130650), Silver-Russell syndrome (SRS; OMIM 180860), Transient Neonatal Diabetes Mellitus (TNDM; OMIM 601410), Kagami-Ogata syndrome (KOS; OMIM 608149), Temple syndrome (TS; OMIM 616222), Prader-Willi syndrome (PWS; OMIM 176270), Angelman syndrome (AS; OMIM 105830) and Pseudohypoparathyroidism Ib (PHP1b; OMIM 103580).
However reporting epigenomic data from clinical molecular tests in laboratory reports or for publication is troubled by the lack of a uniform nomenclature, which is routinely used for DNA sequence variants and mutations. In an effort to clarify the nomenclature, members of the COST action 'European Network for Human Congenital Imprinting Disorders' (BM1208) have suggested names for each DMR. The precise location of each imprinted DMR is based on methyl-seq data from whole blood samples giving base-pair precision. Furthermore, to ensure the same genomic regions are identifiable in different genome builds, all imprinted domains, highlighting imprinted DMRs, have been assigned a Locus Reference Genomic identifier.

Names and locations of imprinted DMRs (table)
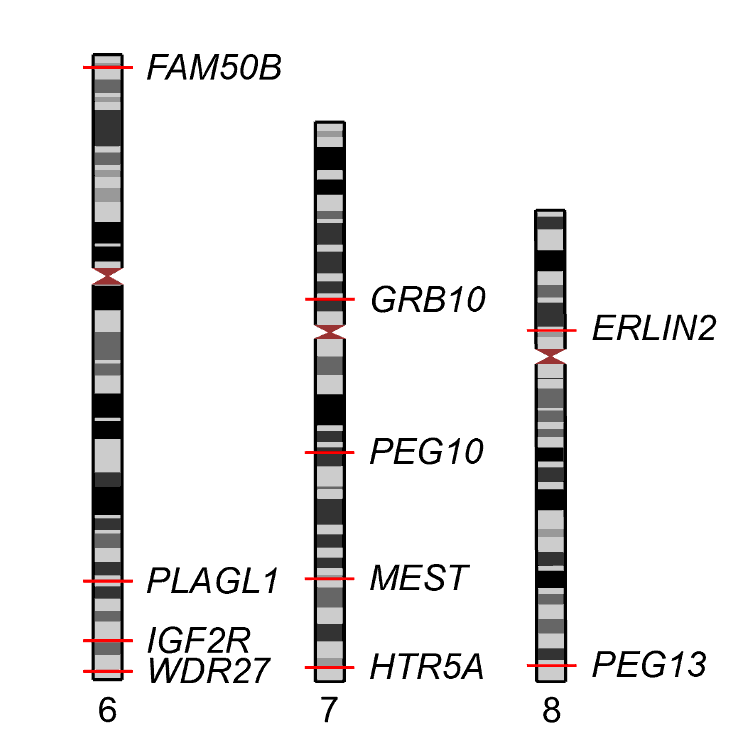
Names and locations of imprinted DMRs (figure)

Probes mapping to imprinted DMRs on the Infinium HumanMethylation450 BeadChip
USEFUL WEBSITES
Links to imprinting related resources.
- UCSC Genome Browser
A specifically designed user friendly webpage for finding gene and transcript sequences, reported SNPs and gene location.
- European Network for Human Congenital Imprinting Disorders
An EU funded network working on diverse research aspects related to imprinting disorders, from clinical diagnosis to molecular characterisation.
- Imprinted Gene Catalogue
A Catalogue of parent-of-origin effects and known/candidate imprinted genes.
- WAMIDEX
Web atlas of murine genomic imprinting, differential expression and epigenetic marks.
- MouseBook Catalogue Imprinting Resource
The original maps of imprinting phenotypes derived from genetics studies in which Robertsonian (Rb) and reciprocal (T) translocations have bee used to generate uniparental disomies and uniparental duplications on whole or selected chromosome regions.
TRAINING OPPORTUNITIES
For graduates and postdoctoral students.
- Graduate: Our lab accepts an average of one new PhD student per year. To be considered, applicants should have a strong background in genetics or developmental biology with first class degree and Masters scores. Candidates should also have good writing/communication skills, excellent teamwork qualities, and a passion for science and research.
- Postdoctoral: We welcome enquiries from postdoctoral applicants and who already have or are competitive for funding awards.